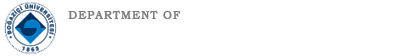NEWS
 |
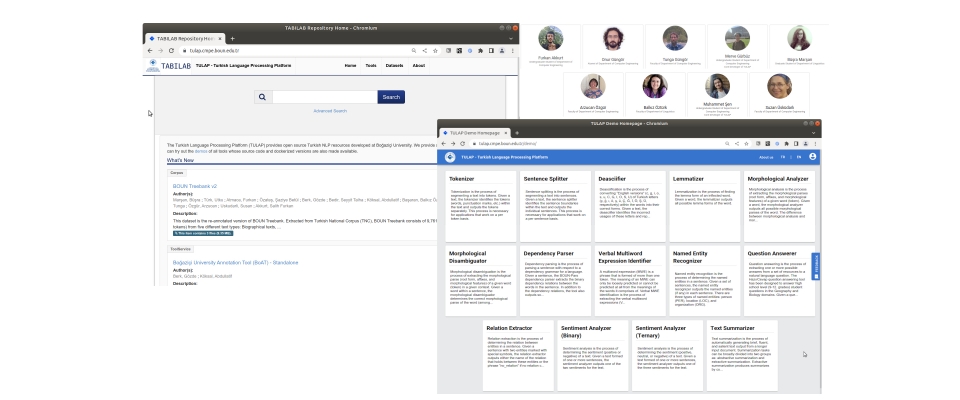
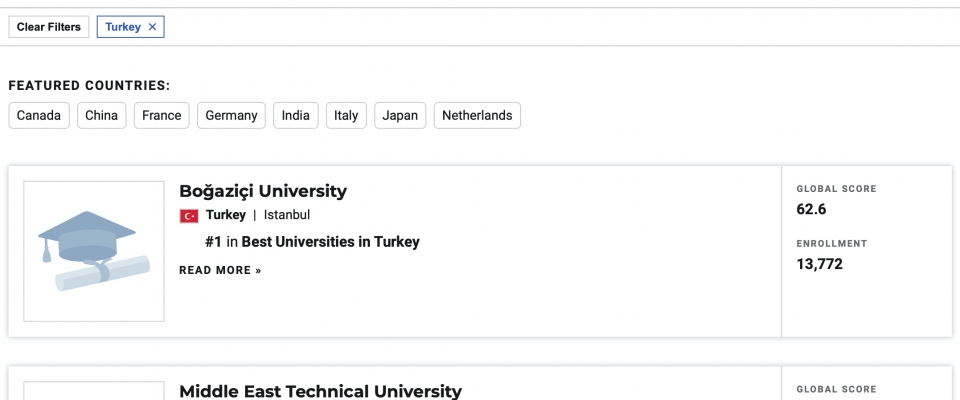
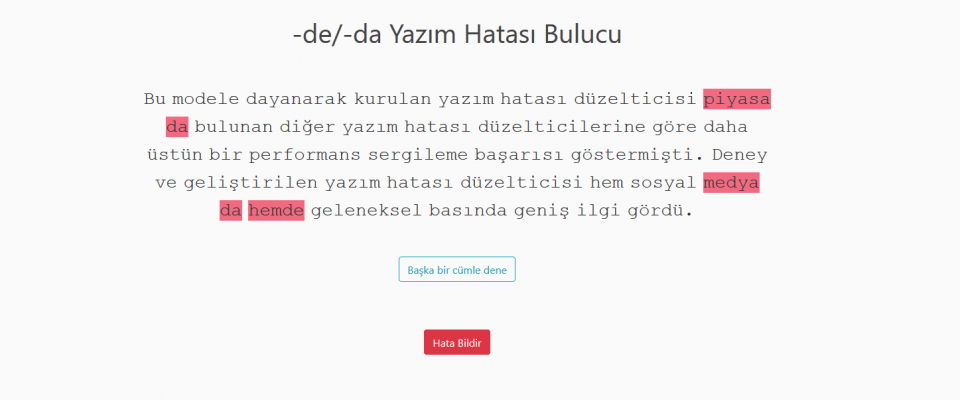
TURNA Türkçe Dil Modeli yayınlandı Boğaziçi Üniversitesi Bilgisayar Mühendisliği Bölümü'ndeki Text Analytics and BIoInformatics Lab (TABILAB) Language Modeling Group proje ekibi TURNA adını verdiği Türkçe dil Read more... |
 |
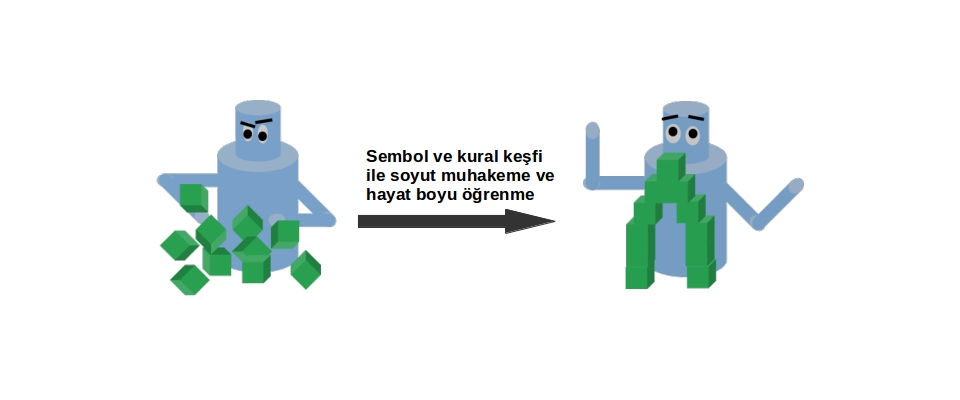
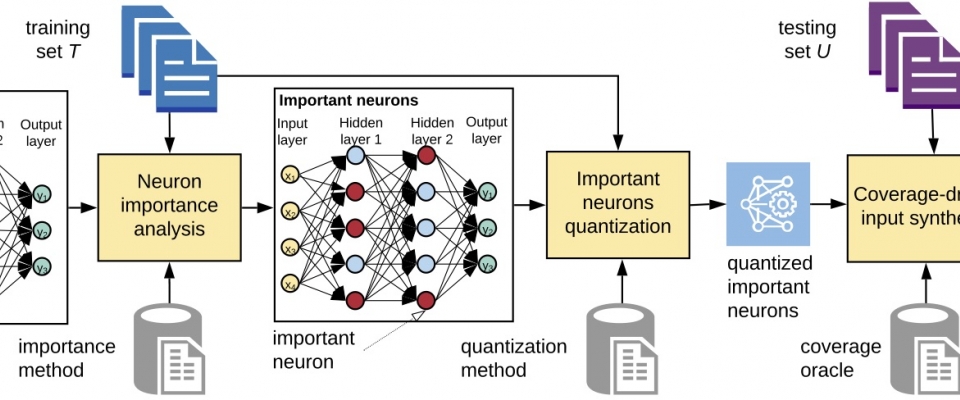
Alper Ahmetoğlu has successfully defended his PhD thesis Title: Neurosymbolic Representations for Lifelong Learning Abstract: This thesis presents a novel framework for robot learning that combines the advantages of deep neural Read more... |
 |
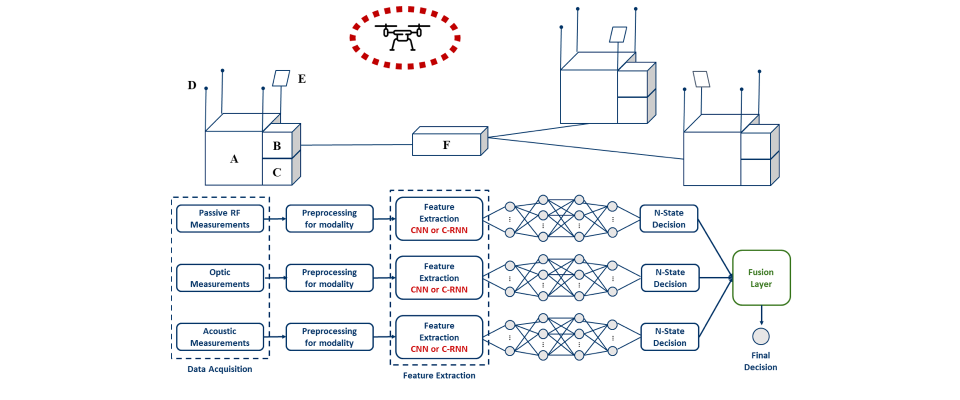
Furkan Oruç has successfully defended his Master's thesis Title: DRONE DETECTION METHODS AND COMPARISON METRICS FOR AUDIO AND VISION MODALITIES Abstract: Drones are unmanned aerial vehicles with a significantly growing Read more... |
 |
Emre Girgin tezini başarıyla sundu Emre Girgin, "Occlusion-aware benchmarking in human pose and shape estimation" başlıklı tezini 8 Ocak 2024'te başarıyla sundu. |
 |
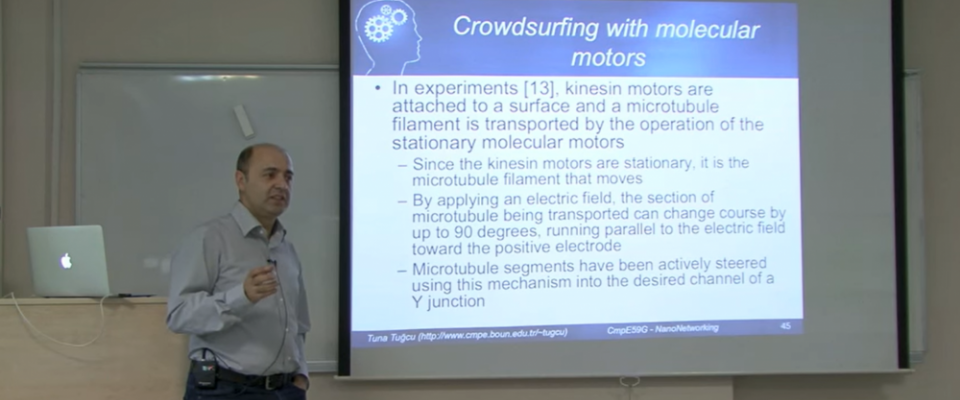
The INVERSE project, in which Assoc. Prof. Dr. Emre Uğur is involved as a partner, has been accepted within the scope of the Horizon Europe program under the CL4-2023-DIGITAL-EMERGING call. Assoc. Prof. Dr. Emre Uğur has received a grant as a partner in the project titled "INteractive robots that intuitiVely lEarn to inVErt tasks by ReaSoning about their Execution Read more... |
Pages
CmpE Events
Please check this page regularly for our upcoming events.
CmpE Announcements
.